BLASTN 2.2.24 [Aug-08-2010]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
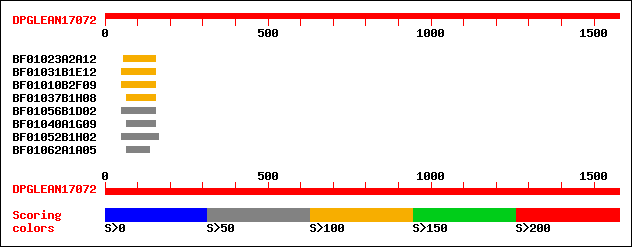
Query= DPGLEAN17072 (UTR included)
(1576 letters)
Database: est
9484 sequences; 8,280,457 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF01023A2A12 137 6e-32
BF01031B1E12 113 9e-25
BF01010B2F09 105 2e-22
BF01037B1H08 103 8e-22
BF01056B1D02 98 5e-20
BF01040A1G09 88 5e-17
BF01052B1H02 84 8e-16
BF01062A1A05 82 3e-15
>BF01023A2A12
Length = 723
Score = 137 bits (69), Expect = 6e-32
Identities = 94/101 (93%), Gaps = 1/101 (0%)
Strand = Plus / Plus
Query: 56 ccaatccgccttgagcgagcgtggtgattaatgctctaactttctccatgtgagaaga-g 114
|||||| ||||||||||||||||||||||||||||||||| |||||||||||| |||| |
Sbjct: 244 ccaatcagccttgagcgagcgtggtgattaatgctctaaccttctccatgtgataagagg 303
Query: 115 cctttactcagcagtggactccgataggctgatgatgatga 155
||||| |||||||||| |||||||||||||||||||||||
Sbjct: 304 cctttgctcagcagtgagctccgataggctgatgatgatga 344
>BF01031B1E12
Length = 730
Score = 113 bits (57), Expect = 9e-25
Identities = 93/105 (88%)
Strand = Plus / Plus
Query: 50 aaattgccaatccgccttgagcgagcgtggtgattaatgctctaactttctccatgtgag 109
|||| ||||| ||| ||||||| ||||||||||||||||||||||| |||||| ||||||
Sbjct: 112 aaatcgccaacccgtcttgagcaagcgtggtgattaatgctctaaccttctccgtgtgag 171
Query: 110 aagagcctttactcagcagtggactccgataggctgatgatgatg 154
||| | ||| ||||||||||| |||||||||||| |||||||||
Sbjct: 172 aaggggatttgctcagcagtgggctccgataggcttatgatgatg 216
>BF01010B2F09
Length = 764
Score = 105 bits (53), Expect = 2e-22
Identities = 95/107 (88%), Gaps = 4/107 (3%)
Strand = Plus / Minus
Query: 50 aaattgccaatccgccttgagcgagcgtggtgattaatgctctaactttctccatgtgag 109
|||| ||||||||| ||||||||||||||||||||||||||||||| |||||||||||||
Sbjct: 366 aaatcgccaatccgtcttgagcgagcgtggtgattaatgctctaaccttctccatgtgag 307
Query: 110 aaga-gcctttactcagcagtggactccgataggctgatgatgatga 155
|||| |||| |||||| | || |||||| |||||| |||||||||
Sbjct: 306 aagacgcct---ctcagcggcgggctccgacaggctggtgatgatga 263
>BF01037B1H08
Length = 758
Score = 103 bits (52), Expect = 8e-22
Identities = 83/92 (90%), Gaps = 1/92 (1%)
Strand = Plus / Plus
Query: 65 cttgagcgagcgtggtgattaatgctctaactttctccatgtgagaaga-gcctttactc 123
||||||| ||||||||| ||||||||||||| |||| |||||||||||| |||||| |||
Sbjct: 257 cttgagcaagcgtggtgcttaatgctctaaccttctacatgtgagaagaggcctttgctc 316
Query: 124 agcagtggactccgataggctgatgatgatga 155
||| |||| |||||||||||||||| ||||||
Sbjct: 317 agctgtgggctccgataggctgatggtgatga 348
>BF01056B1D02
Length = 778
Score = 97.6 bits (49), Expect = 5e-20
Identities = 94/107 (87%), Gaps = 4/107 (3%)
Strand = Plus / Plus
Query: 50 aaattgccaatccgccttgagcgagcgtggtgattaatgctctaactttctccatgtgag 109
|||| ||||| ||| ||||||| ||||||||| ||||||||||||| ||| |||||||
Sbjct: 639 aaatcgccaacccgtcttgagcaagcgtggtggttaatgctctaaccttc---atgtgag 695
Query: 110 aagag-cctttactcagcagtggactccgataggctgatgatgatga 155
||||| ||||| ||||||||||| |||||||||||||||||| ||||
Sbjct: 696 aagaggcctttgctcagcagtgggctccgataggctgatgataatga 742
>BF01040A1G09
Length = 753
Score = 87.7 bits (44), Expect = 5e-17
Identities = 81/92 (88%), Gaps = 1/92 (1%)
Strand = Plus / Plus
Query: 65 cttgagcgagcgtggtgattaatgctctaactttctccatgtgagaagagcctt-tactc 123
||||||| ||||||| |||||||||| ||| ||||||||| |||||||| ||| | |||
Sbjct: 45 cttgagcaagcgtggcgattaatgcttaaaccttctccatgcgagaagaggcttatgctc 104
Query: 124 agcagtggactccgataggctgatgatgatga 155
|||||||| ||||||||||||||||||||||
Sbjct: 105 agcagtggcttccgataggctgatgatgatga 136
>BF01052B1H02
Length = 790
Score = 83.8 bits (42), Expect = 8e-16
Identities = 101/118 (85%), Gaps = 2/118 (1%)
Strand = Plus / Minus
Query: 50 aaattgccaatccgccttgagcgagcgtggtgattaatgctctaactttctccatgtgag 109
|||| ||||| ||| ||||||| ||||||||| ||||||||||||||||||||||| |||
Sbjct: 373 aaatcgccaacccgtcttgagcaagcgtggtgcttaatgctctaactttctccatgggag 314
Query: 110 aaga-gcctttactcagcagtggactccgataggctgatgatgatga-gatgatgatg 165
|| | || ||| |||| |||||| ||| ||| |||| |||||| ||| ||||||||||
Sbjct: 313 aaaaggcgtttgctcaacagtgggctctgatgggctaatgatggtgatgatgatgatg 256
>BF01062A1A05
Length = 777
Score = 81.8 bits (41), Expect = 3e-15
Identities = 66/73 (90%), Gaps = 1/73 (1%)
Strand = Plus / Plus
Query: 65 cttgagcgagcgtggtgattaatgctctaactttctccatgtgagaaga-gcctttactc 123
||||||| ||||||||||||||||||||||| |||||| |||||||||| |||||| ||
Sbjct: 514 cttgagcaagcgtggtgattaatgctctaaccttctccttgtgagaagaggcctttgctt 573
Query: 124 agcagtggactcc 136
|||||||| ||||
Sbjct: 574 agcagtgggctcc 586
Database: est
Posted date: Jul 14, 2011 2:25 PM
Number of letters in database: 8,280,457
Number of sequences in database: 9484
Lambda K H
1.37 0.711 1.31
Gapped
Lambda K H
1.37 0.711 1.31
Matrix: blastn matrix:1 -3
Gap Penalties: Existence: 5, Extension: 2
Number of Sequences: 9484
Number of Hits to DB: 179,108
Number of extensions: 18273
Number of successful extensions: 18273
Number of sequences better than 1.0e-10: 8
Number of HSP's gapped: 18268
Number of HSP's successfully gapped: 8
Length of query: 1576
Length of database: 8,280,457
Length adjustment: 17
Effective length of query: 1559
Effective length of database: 8,119,229
Effective search space: 12657878011
Effective search space used: 12657878011
X1: 11 (21.8 bits)
X2: 15 (29.7 bits)
X3: 50 (99.1 bits)
S1: 12 (24.3 bits)
S2: 34 (67.9 bits)

