BLASTN 2.2.24 [Aug-08-2010]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
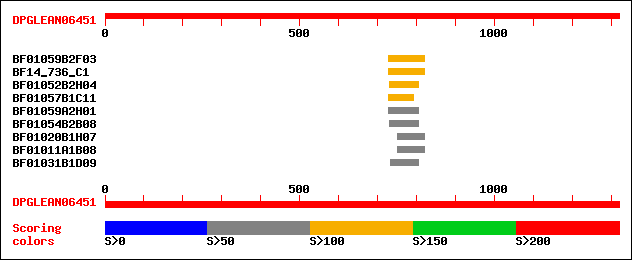
Query= DPGLEAN06451 (UTR included)
(1318 letters)
Database: est
9484 sequences; 8,280,457 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF01059B2F03 125 2e-28
BF14_736_C1 125 2e-28
BF01052B2H04 121 3e-27
BF01057B1C11 109 1e-23
BF01059A2H01 100 1e-20
BF01054B2B08 82 3e-15
BF01020B1H07 72 2e-12
BF01011A1B08 72 2e-12
BF01031B1D09 68 4e-11
>BF01059B2F03
Length = 703
Score = 125 bits (63), Expect = 2e-28
Identities = 84/91 (92%)
Strand = Plus / Minus
Query: 728 ggtagagcctagctgtagtcatgtaactacagatcgaacatgggctggctcgaccgggga 787
||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||
Sbjct: 127 ggtagagcctagctgtagtcatgtaactacagatcgaacatgggctggctcgagcgggga 68
Query: 788 agtaccaccctctcacagtaggttggcgtga 818
| ||||||| ||||||| || | |||||||
Sbjct: 67 atgaccaccccctcacagaagatcggcgtga 37
>BF14_736_C1
Length = 796
Score = 125 bits (63), Expect = 2e-28
Identities = 84/91 (92%)
Strand = Plus / Plus
Query: 728 ggtagagcctagctgtagtcatgtaactacagatcgaacatgggctggctcgaccgggga 787
|||||||||||||||| |||| || ||||||| |||||||||||||||||||||||||||
Sbjct: 586 ggtagagcctagctgtggtcaagttactacagctcgaacatgggctggctcgaccgggga 645
Query: 788 agtaccaccctctcacagtaggttggcgtga 818
|||||||||||||||||| || | |||||||
Sbjct: 646 agtaccaccctctcacagaagatcggcgtga 676
>BF01052B2H04
Length = 668
Score = 121 bits (61), Expect = 3e-27
Identities = 73/77 (94%)
Strand = Plus / Minus
Query: 729 gtagagcctagctgtagtcatgtaactacagatcgaacatgggctggctcgaccggggaa 788
||||||||| ||||| ||||||||||||||| ||||||||||||||||||| ||||||||
Sbjct: 470 gtagagcctggctgtggtcatgtaactacagctcgaacatgggctggctcgtccggggaa 411
Query: 789 gtaccaccctctcacag 805
|||||||||||||||||
Sbjct: 410 gtaccaccctctcacag 394
>BF01057B1C11
Length = 848
Score = 109 bits (55), Expect = 1e-23
Identities = 61/63 (96%)
Strand = Plus / Plus
Query: 728 ggtagagcctagctgtagtcatgtaactacagatcgaacatgggctggctcgaccgggga 787
|||||||||||||||| ||||||||||||||||||||||||||||||||||| |||||||
Sbjct: 51 ggtagagcctagctgtggtcatgtaactacagatcgaacatgggctggctcgtccgggga 110
Query: 788 agt 790
|||
Sbjct: 111 agt 113
>BF01059A2H01
Length = 865
Score = 99.6 bits (50), Expect = 1e-20
Identities = 71/78 (91%)
Strand = Plus / Minus
Query: 728 ggtagagcctagctgtagtcatgtaactacagatcgaacatgggctggctcgaccgggga 787
|||||||||||||||| |||| |||||||||| || |||||| ||||||| ||||| |||
Sbjct: 348 ggtagagcctagctgtggtcacgtaactacagctccaacatgagctggcttgaccgtgga 289
Query: 788 agtaccaccctctcacag 805
||||||||||||||||||
Sbjct: 288 agtaccaccctctcacag 271
>BF01054B2B08
Length = 807
Score = 81.8 bits (41), Expect = 3e-15
Identities = 69/77 (89%), Gaps = 1/77 (1%)
Strand = Plus / Plus
Query: 730 tagagcctagctgta-gtcatgtaactacagatcgaacatgggctggctcgaccggggaa 788
||||||||||||||| |||| |||||||||| ||||||| ||| |||||||||||||||
Sbjct: 581 tagagcctagctgtatgtcaagtaactacagctcgaacaagggttggctcgaccggggat 640
Query: 789 gtaccaccctctcacag 805
|||| | ||||||||||
Sbjct: 641 gtactagcctctcacag 657
>BF01020B1H07
Length = 733
Score = 71.9 bits (36), Expect = 2e-12
Identities = 60/68 (88%)
Strand = Plus / Minus
Query: 751 taactacagatcgaacatgggctggctcgaccggggaagtaccaccctctcacagtaggt 810
||||||||| || |||||||||||||||||| ||| |||||||| |||||||||| || |
Sbjct: 115 taactacagctcaaacatgggctggctcgacggggcaagtaccatcctctcacagaagat 56
Query: 811 tggcgtga 818
|||||||
Sbjct: 55 cggcgtga 48
>BF01011A1B08
Length = 853
Score = 71.9 bits (36), Expect = 2e-12
Identities = 60/68 (88%)
Strand = Plus / Minus
Query: 751 taactacagatcgaacatgggctggctcgaccggggaagtaccaccctctcacagtaggt 810
||||||||| || |||||||||||||||||| ||| |||||||| |||||||||| || |
Sbjct: 153 taactacagctcaaacatgggctggctcgacggggcaagtaccatcctctcacagaagat 94
Query: 811 tggcgtga 818
|||||||
Sbjct: 93 cggcgtga 86
>BF01031B1D09
Length = 754
Score = 67.9 bits (34), Expect = 4e-11
Identities = 64/74 (86%)
Strand = Plus / Plus
Query: 732 gagcctagctgtagtcatgtaactacagatcgaacatgggctggctcgaccggggaagta 791
|||||||| ||| |||| || ||| ||| ||||||| ||||||||| ||| ||| |||||
Sbjct: 476 gagcctagttgtggtcaagttactgcagctcgaacaagggctggcttgacggggcaagta 535
Query: 792 ccaccctctcacag 805
||||||||||||||
Sbjct: 536 ccaccctctcacag 549
Database: est
Posted date: Jul 14, 2011 2:25 PM
Number of letters in database: 8,280,457
Number of sequences in database: 9484
Lambda K H
1.37 0.711 1.31
Gapped
Lambda K H
1.37 0.711 1.31
Matrix: blastn matrix:1 -3
Gap Penalties: Existence: 5, Extension: 2
Number of Sequences: 9484
Number of Hits to DB: 228,854
Number of extensions: 6700
Number of successful extensions: 6700
Number of sequences better than 1.0e-10: 9
Number of HSP's gapped: 6699
Number of HSP's successfully gapped: 9
Length of query: 1318
Length of database: 8,280,457
Length adjustment: 17
Effective length of query: 1301
Effective length of database: 8,119,229
Effective search space: 10563116929
Effective search space used: 10563116929
X1: 11 (21.8 bits)
X2: 15 (29.7 bits)
X3: 50 (99.1 bits)
S1: 12 (24.3 bits)
S2: 34 (67.9 bits)

